Audiology of the Future with Next Generation, Whole Human Genome Sequencing
Renowned auditory researcher Dr. Robert Harrison brings us up to date on information and research from the Labs. Appropriately titled “From the Labs to the Clinics”, Bob is involved in laboratory and applied/clinical research, including evoked potential and otoacoustic emission studies and behavioural studies of speech and language development in children with cochlear implants. For a little insight into Bob’s interests outside the lab and the clinic, we invite you to climb aboard Bob’s Garden Railway.
In my next few columns, I will explore new and exciting studies on using whole human genome sequencing as an emerging clinical tool for audiology. Imagine 20–30 years into the future when you might routinely order a whole genome sequence of a client, to provide an accurate diagnosis of a hearing or vestibular problem and guide optimal intervention. This will be the ultimate “personalized medicine” we hear so much about. I am convinced that the scenario above will come to pass. There are already many potential applications of whole genome sequencing in audiological areas such as neonatal screening, diagnosis of genetic causes of hearing loss, and determining risk factors (genetic disposition) for age-related hearing problems. I will discuss the current science around these and other audiological applications in future editions of Canadian Audiologist. But first, I want to get you up to speed today with a short overview of this exciting, emerging field.


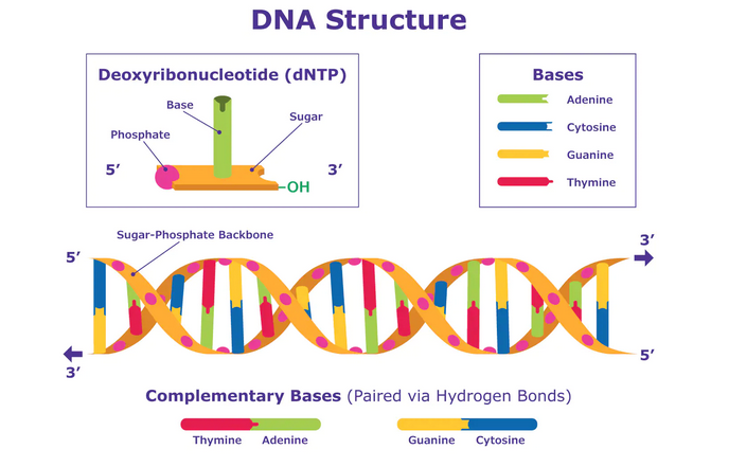
The original methodology for whole genome sequencing was invented by Frederick Sanger in 1975, and originally applied to sequencing bacterial DNA.1,2 This technique is usually referred to as Sanger sequencing.
The total cost of generating the first human genome sequence3 was many billions of dollars (yes, billions!). The US funding contribution alone was $3 billion. Within a decade, and with improved technology, the cost of an individual, high-quality genome sequence fell to $14 million by 2006. Ten years later (2016) the cost was down to a few thousand $, but data processing times were lengthy - days to weeks even with highly automated methods. More recently, “new generation sequencing” methods and shortcuts have been developed to reduce time and costs. For example, pieces of DNA (exons) that provide instructions for making proteins make up a small percentage of the whole genome and can be sequenced rapidly. It is thought that most (but not all) mutations that cause disease occur in exons, so whole exome sequencing can efficiently identify mutations that cause disease. Today, such whole exome sequencing costs a few hundred dollars and takes about 5 hours. These costs/times now make whole genome sequencing an affordable research tool and convenient for potential clinical applications.
There is no doubt that the ability to map the human genome has the promise of a new era in healthcare. The earliest clinical applications were in detecting unknown, disease-causing gene mutations. In some cases, identification of the genes (genotype) and their biological function (phenotype) has led to successful treatment to restore normal function. Our accumulation of knowledge about the genes mutations responsible for various diseases has provided us with gene panels that can be used in genetic screening for individual patients. We are all familiar with certain types of congenital, genetically related hearing problems in audiology, and we presently use genetic screening panels to identify known gene mutations. However, hundreds (if not thousands) of rare and unknown gene mutations (single or multiple) are linked to hearing loss; these can be potentially revealed by whole genome sequencing.
With the potential low cost and availability of whole genome mapping, we might well see this used for UNIVERSAL neonatal screening to complement our hearing screening.4,5 This will more immediately help to define the etiology of a hearing problem and inform optimal intervention. Further to genome mapping for congenital hearing loss, we could move to whole genome mapping in later life to determine genetic risk factors for developing hearing loss, tinnitus, or balance problems related to noise exposure or aging.6-9 Regarding another audiology application, I recently heard a conference presentation on whole genome sequencing of a large cohort of patients with Meniere’s disease. As we know, this grouping is very much a “mixed bag,” and the study's intent was to define genetic subtypes with distinctly different etiology and pathological features. Such a sub-classification could be useful to finally understand the causes of the disease and define treatment options. Perhaps the same methods can be used to subdivide the spectrum of disease in ANSD.Clearly, human genome sequencing has much to offer the field of audiology. In future columns we will further discuss these potential applications.
References
- Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes JC, Hutchison CA, Slocombe PM, Smith M. (1977). Nucleotide sequence of bacteriophage φX174 DNA. Nature. 1977 Feb;265 (5596):687-95.
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proceedings of the national academy of sciences. 1977 Dec;74(12):5463-7. https://doi.org/10.1073/pnas.74.12.546
- International Human Genome Sequencing Consortium. (2004). Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 (2004). https://doi.org/10.1038/nature03001
- 4. Tutty, E., Amor, D. J., Jarmolowicz, A., Paton, K., & Downie, L. (2021). Personal utility of genomic sequencing for infants with congenital deafness. American Journal of Medical Genetics Part A, 185A: 3634– 3643. https://doi.org/10.1002/ajmg.a.62411
- Audrey C. Woerne, Renata C. Gallagher, Jerry Vockley, Aashish N. Adhikari. (2021). The Use of Whole Genome and Exome Sequencing for Newborn Screening: Challenges and Opportunities for Population Health Front. Pediatr., 19 July 2021 https://doi.org/10.3389/fped.2021.663752
- Ivarsdottir, E.V., Holm, H., Benonisdottir, S. et al. (2021). The genetic architecture of age-related hearing impairment revealed by genome-wide association analysis. Commun Biol 4, 706 (2021). https://doi.org/10.1038/s42003-021-02224-9
- Nisenbaum, Eric; Prentiss, Sandra; Yan, Denise; Nourbakhsh, Aida; Smeal, Molly; Holcomb et al. (2021). Screening Strategies for Deafness Genes and Functional Outcomes in Cochlear Implant Patients. Otology & Neurotology: January 2021 - Volume 42 - Issue 1 - p 180-187. https://doi:10.1097/MAO.0000000000002969
- Praveen, K., Dobbyn, L., Gurski, L. et al. (2022) Population-scale analysis of common and rare genetic variation associated with hearing loss in adults. Commun Biol 5, 540 (2022). https://doi.org/10.1038/s42003-022-03408-7
- Molina-Ramírez, LP, Burkitt-Wright, EMM, Saeed, H, et al. (2021). The diagnostic utility of clinical exome sequencing in 60 patients with hearing loss disorders: A single-institution experience. Clin Otolaryngol. 2021; 46: 1257– 1262. https://doi.org/10.1111/coa.13826


